BIOSENSORS, MOTOR PROTEINS AND MORE
MARTIN WEBB
SCIENCE
Home > Research topics > Helicases
HELICASES
INTRODUCTION
DNA helicases are ubiquitous motor proteins that couple the free energy of ATP hydrolysis to the separation of double-stranded DNA into its component single strands.
They are involved in a wide range of DNA processing, including replication and repair, but often occur as part of larger protein complexes that are multifunctional with coordinated processing of the DNA.
We have studied several helicases with different biological functions and from different subfamilies. The biochemical mechanisms were investigated, by which such important DNA processing occurs and may be controlled. This relates coupling of the ATPase cycle to structural and functional changes that lead to movement of the protein along the DNA.
I would like to acknowledge and thank all the people from my lab and elsewhere, who contributed to this work. Their names are on the cited publications.
HELICASE PcrA
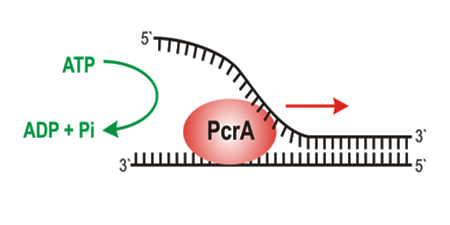
Key measurements were on the prokaryotic helicase PcrA, as an exemplar of helicases in general. This helicase is monomeroic and one of its functions is in rolling circle replication, described below.
On its own, it has rather poor helicase (unwinding) activity, but rapidly translocates along ssDNA.
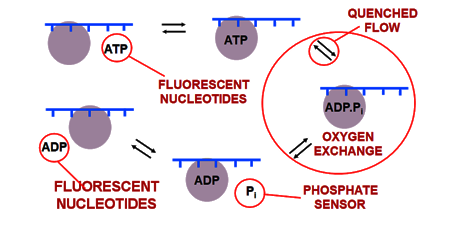
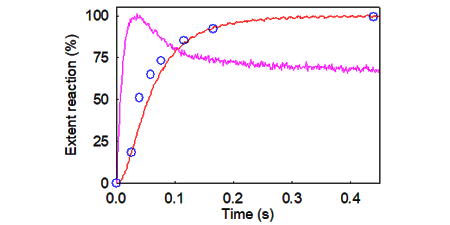
Some key measurements were on PcrA alone and its interactions with DNA. This included developing methods to measure translocation and also elucidating the ATPase cycle to determine which key steps in that cycle couple to movement.
This movement was shown to be strictly coupled to ATP hydrolysis at one ATP per base
ROLLING CIRCLE PLASMID REPLICATION
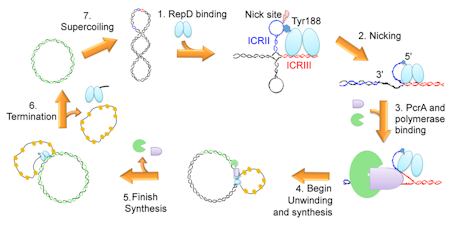
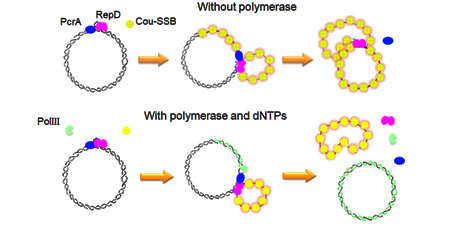
PcrA is involved in replication of certain plasmids in gram-positive bacteria, in particular those promiscuous plasmids containing antibiotic resistance genes. As such, this system is a potential target for drug discovery, particularly as the rolling-circle replication mechanism differs significantly from the standard, synchronized replication of genomic DNA.
The double-stranded origin of replication is recognized by an initiator protein (RepD), which nicks and covalently attaches to one strand. PcrA and a polymerase are recruited at this origin of replication. Subsequent translocation through several thousand base-pairs of DNA gives complete separation of parent strands and replication of one of them.
Apart from PcrA, only the initiator protein, RepD, and a polymerase are required, so this is attractive to study a complete replication machine.
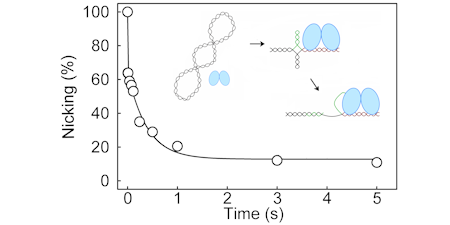
INITIATOR PROTEIN RepD
The initiator protein, RepD, acting as a homodimer, is responsible for nicking one strand at the origin, so enabling the helicase and p[olymerase to bind and begin their functions.
It alone can enable PcrA to unwind thousands of dsDNA bases. Finally, it is responsible for terminating replication of the first parent strand, by resolving the double-length DNA into two circles, producing a daughter dsDNA plasmid and a ssDNA plasmid. The latter is replicated by a different polymerase.
We have determined the mechanism by which RepD nicks the DNA and examined how it greatly increases PcrA processivity.
EXTENDING STUDIES
In collaboration with Justin Molloy (Francis Crick Institute) we have used TIRF to measure unwinding of single plasmids. A wide variation in unwinding rate produces a similar rate to that seen in bulk measurements, but single molecules show pauses. This suggests that the unwinding may vary with the physical state of individual plasmids, such as degree of supercoiling. (Key refs: 106, 120)
Also with Justin Molloy, magnetic tweezers were used to control the degree of supercoiling and show how RepD activity is tightly controlled by supercoiling through cruciform formation at the nick site, ensuring that only intact plasmids get activated for replication. (130)